




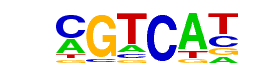

















| p-value: | 1e-1208 |
| log p-value: | -2.782e+03 |
| Information Content per bp: | 1.921 |
| Number of Target Sequences with motif | 6922.0 |
| Percentage of Target Sequences with motif | 42.16% |
| Number of Background Sequences with motif | 5374.8 |
| Percentage of Background Sequences with motif | 17.28% |
| Average Position of motif in Targets | 202.9 +/- 84.2bp |
| Average Position of motif in Background | 193.1 +/- 141.3bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.31 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
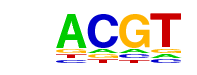
| 1 | 0.941 |  | 1e-1064 | -2450.537369 | 55.62% | 29.37% | motif file (matrix) |
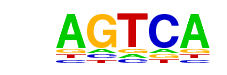
| 2 | 0.849 |  | 1e-985 | -2268.221661 | 39.10% | 16.96% | motif file (matrix) |
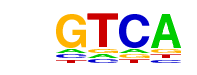
| 3 | 0.970 |  | 1e-983 | -2265.526937 | 23.67% | 7.00% | motif file (matrix) |
| 4 | 0.977 |  | 1e-974 | -2244.857388 | 22.35% | 6.33% | motif file (matrix) |
| 5 | 0.652 |  | 1e-946 | -2179.472047 | 22.62% | 6.62% | motif file (matrix) |
| 6 | 0.855 |  | 1e-898 | -2069.631290 | 67.45% | 42.56% | motif file (matrix) |
| 7 | 0.768 |  | 1e-558 | -1285.035614 | 27.50% | 12.69% | motif file (matrix) |
| 8 | 0.725 |  | 1e-482 | -1109.886322 | 46.37% | 28.98% | motif file (matrix) |
| 9 | 0.661 |  | 1e-469 | -1080.459994 | 72.87% | 55.30% | motif file (matrix) |
| 10 | 0.779 |  | 1e-467 | -1076.858432 | 73.87% | 56.43% | motif file (matrix) |
| 11 | 0.616 |  | 1e-450 | -1037.871636 | 46.85% | 29.94% | motif file (matrix) |
| 12 | 0.711 |  | 1e-407 | -938.875532 | 45.50% | 29.50% | motif file (matrix) |
| 13 | 0.808 |  | 1e-374 | -861.984067 | 53.26% | 37.32% | motif file (matrix) |
| 14 | 0.811 |  | 1e-343 | -791.429616 | 13.87% | 5.53% | motif file (matrix) |
| 15 | 0.616 |  | 1e-262 | -605.297066 | 46.26% | 33.19% | motif file (matrix) |
| 16 | 0.644 |  | 1e-243 | -559.690305 | 64.64% | 51.75% | motif file (matrix) |
| 17 | 0.648 |  | 1e-198 | -457.981492 | 77.82% | 67.15% | motif file (matrix) |
| 18 | 0.685 |  | 1e-175 | -404.710029 | 56.94% | 45.91% | motif file (matrix) |
| 19 | 0.837 |  | 1e-127 | -294.686833 | 94.46% | 89.09% | motif file (matrix) |
| 20 | 0.732 |  | 1e-94 | -216.945985 | 14.20% | 9.21% | motif file (matrix) |
| 21 | 0.656 |  | 1e-62 | -143.942009 | 10.89% | 7.27% | motif file (matrix) |
| 22 | 0.718 |  | 1e-61 | -140.829028 | 6.94% | 4.13% | motif file (matrix) |
| 23 | 0.605 |  | 1e-59 | -136.401771 | 79.48% | 74.04% | motif file (matrix) |
| 24 | 0.729 |  | 1e-34 | -78.885167 | 17.77% | 14.31% | motif file (matrix) |