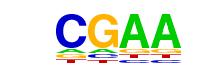
PB0034.1_Irf4_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.73
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CGAA------
CGTATCGAAACCAAA |
|


|
|
PB0035.1_Irf5_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.72
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------CGAA-----
ATAAACCGAAACCAA |
|


|
|
POL008.1_DCE_S_I/Jaspar
| Match Rank: | 3 |
| Score: | 0.70
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CGAA--
NGAAGC |
|


|
|
PB0036.1_Irf6_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----CGAA--------
CTGATCGAAACCAAAGT |
|


|
|
PB0037.1_Isgf3g_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.66
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------CGAA-----
CAAAATCGAAACTAA |
|


|
|
IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CGAA----
ACTGAAACCA |
|


|
|
PB0033.1_Irf3_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.60
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------CGAA----
GAGAACCGAAACTG |
|


|
|
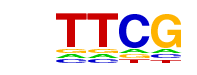
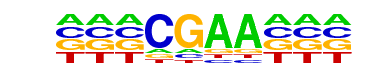
NFATC1/MA0624.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CGAA---
NNTGGAAANN |
|

|
|
SPIB/MA0081.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.60
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CGAA
AGAGGAA |
|


|
|
CENPB/MA0637.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.60
| | Offset: | -11
| | Orientation: | forward strand |
| Alignment: | -----------CGAA
CCCGCATACAACGAA |
|


|
|