








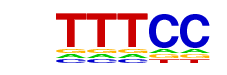




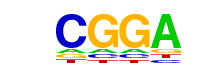



| p-value: | 1e-400 |
| log p-value: | -9.224e+02 |
| Information Content per bp: | 1.889 |
| Number of Target Sequences with motif | 8311.0 |
| Percentage of Target Sequences with motif | 50.62% |
| Number of Background Sequences with motif | 10674.9 |
| Percentage of Background Sequences with motif | 34.31% |
| Average Position of motif in Targets | 201.5 +/- 106.9bp |
| Average Position of motif in Background | 197.8 +/- 143.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.75 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.674 |  | 1e-324 | -746.720774 | 28.01% | 16.06% | motif file (matrix) |
| 2 | 0.935 |  | 1e-281 | -647.839244 | 38.28% | 25.52% | motif file (matrix) |
| 3 | 0.666 |  | 1e-244 | -561.896516 | 50.93% | 38.08% | motif file (matrix) |
| 4 | 0.978 |  | 1e-231 | -532.461918 | 38.94% | 27.23% | motif file (matrix) |
| 5 | 0.709 |  | 1e-222 | -511.528373 | 41.43% | 29.72% | motif file (matrix) |
| 6 | 0.756 |  | 1e-221 | -509.353761 | 31.92% | 21.25% | motif file (matrix) |
| 7 | 0.699 |  | 1e-191 | -440.281113 | 41.76% | 30.82% | motif file (matrix) |
| 8 | 0.735 |  | 1e-153 | -353.107964 | 55.75% | 45.46% | motif file (matrix) |
| 9 | 0.687 |  | 1e-143 | -330.582961 | 70.25% | 60.69% | motif file (matrix) |
| 10 | 0.768 |  | 1e-125 | -288.672722 | 74.92% | 66.33% | motif file (matrix) |
| 11 | 0.710 |  | 1e-124 | -286.009069 | 47.91% | 38.78% | motif file (matrix) |
| 12 | 0.665 |  | 1e-116 | -268.667804 | 38.67% | 30.23% | motif file (matrix) |
| 13 | 0.680 |  | 1e-83 | -191.534606 | 65.77% | 58.40% | motif file (matrix) |
| 14 | 0.627 |  | 1e-57 | -132.915140 | 49.94% | 43.69% | motif file (matrix) |
| 15 | 0.873 |  | 1e-37 | -85.414881 | 67.18% | 62.38% | motif file (matrix) |
| 16 | 0.621 |  | 1e-22 | -52.674456 | 5.45% | 3.87% | motif file (matrix) |