

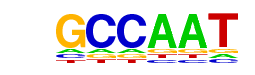






| p-value: | 1e-244 |
| log p-value: | -5.633e+02 |
| Information Content per bp: | 1.627 |
| Number of Target Sequences with motif | 4024.0 |
| Percentage of Target Sequences with motif | 24.51% |
| Number of Background Sequences with motif | 4534.4 |
| Percentage of Background Sequences with motif | 14.58% |
| Average Position of motif in Targets | 201.1 +/- 102.9bp |
| Average Position of motif in Background | 191.2 +/- 128.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.37 |
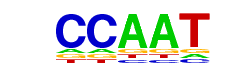
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.683 |  | 1e-202 | -467.138029 | 45.36% | 33.87% | motif file (matrix) |
| 2 | 0.945 |  | 1e-180 | -416.658323 | 22.22% | 13.91% | motif file (matrix) |
| 3 | 0.937 |  | 1e-157 | -362.011210 | 18.62% | 11.46% | motif file (matrix) |
| 4 | 0.619 |  | 1e-124 | -287.295890 | 55.31% | 46.03% | motif file (matrix) |
| 5 | 0.896 |  | 1e-110 | -255.179190 | 44.12% | 35.62% | motif file (matrix) |
| 6 | 0.696 |  | 1e-106 | -245.499209 | 22.43% | 15.85% | motif file (matrix) |
| 7 | 0.725 |  | 1e-82 | -190.104606 | 26.12% | 19.90% | motif file (matrix) |
| 8 | 0.809 |  | 1e-59 | -137.975150 | 15.99% | 11.69% | motif file (matrix) |