
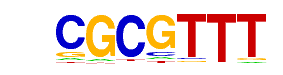
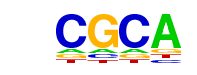








| p-value: | 1e-191 |
| log p-value: | -4.420e+02 |
| Information Content per bp: | 1.554 |
| Number of Target Sequences with motif | 11151.0 |
| Percentage of Target Sequences with motif | 67.92% |
| Number of Background Sequences with motif | 17620.8 |
| Percentage of Background Sequences with motif | 56.64% |
| Average Position of motif in Targets | 201.5 +/- 112.6bp |
| Average Position of motif in Background | 197.2 +/- 137.2bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.53 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.736 |  | 1e-160 | -368.858578 | 59.96% | 49.45% | motif file (matrix) |
| 2 | 0.820 |  | 1e-156 | -361.452650 | 54.59% | 44.18% | motif file (matrix) |
| 3 | 0.702 |  | 1e-151 | -348.205672 | 70.77% | 60.97% | motif file (matrix) |
| 4 | 0.630 |  | 1e-111 | -257.814190 | 49.16% | 40.46% | motif file (matrix) |
| 5 | 0.602 |  | 1e-92 | -213.261806 | 22.81% | 16.60% | motif file (matrix) |
| 6 | 0.806 |  | 1e-90 | -207.892338 | 41.95% | 34.35% | motif file (matrix) |
| 7 | 0.792 |  | 1e-84 | -195.030326 | 61.95% | 54.39% | motif file (matrix) |
| 8 | 0.835 |  | 1e-76 | -177.219693 | 48.74% | 41.53% | motif file (matrix) |
| 9 | 0.622 |  | 1e-55 | -127.555150 | 69.32% | 63.48% | motif file (matrix) |