

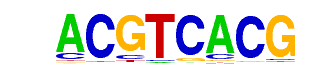
















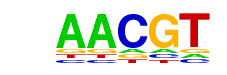

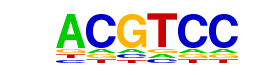


| p-value: | 1e-78 |
| log p-value: | -1.818e+02 |
| Information Content per bp: | 1.768 |
| Number of Target Sequences with motif | 2382.0 |
| Percentage of Target Sequences with motif | 50.83% |
| Number of Background Sequences with motif | 14741.1 |
| Percentage of Background Sequences with motif | 36.49% |
| Average Position of motif in Targets | 969.6 +/- 416.4bp |
| Average Position of motif in Background | 1005.5 +/- 578.7bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.44 |
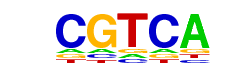
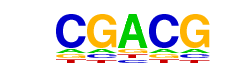
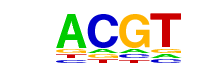
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.954 |  | 1e-64 | -148.704086 | 48.12% | 35.27% | motif file (matrix) |
| 2 | 0.933 |  | 1e-62 | -144.891608 | 45.01% | 32.50% | motif file (matrix) |
| 3 | 0.805 |  | 1e-48 | -111.734029 | 34.83% | 24.61% | motif file (matrix) |
| 4 | 0.881 |  | 1e-47 | -110.126781 | 81.16% | 71.42% | motif file (matrix) |
| 5 | 0.740 |  | 1e-38 | -89.682385 | 53.29% | 43.17% | motif file (matrix) |
| 6 | 0.629 |  | 1e-32 | -75.976568 | 65.04% | 55.91% | motif file (matrix) |
| 7 | 0.705 |  | 1e-31 | -73.256510 | 56.02% | 46.89% | motif file (matrix) |
| 8 | 0.669 |  | 1e-29 | -68.028864 | 93.62% | 88.46% | motif file (matrix) |
| 9 | 0.635 |  | 1e-29 | -67.384782 | 80.20% | 72.68% | motif file (matrix) |
| 10 | 0.646 |  | 1e-29 | -66.944133 | 64.77% | 56.22% | motif file (matrix) |
| 11 | 0.749 |  | 1e-28 | -65.847113 | 61.08% | 52.50% | motif file (matrix) |
| 12 | 0.601 |  | 1e-27 | -64.349163 | 60.29% | 51.80% | motif file (matrix) |
| 13 | 0.662 |  | 1e-26 | -61.492305 | 34.27% | 26.68% | motif file (matrix) |
| 14 | 0.649 |  | 1e-23 | -53.064110 | 75.10% | 68.08% | motif file (matrix) |
| 15 | 0.826 |  | 1e-22 | -52.169173 | 92.40% | 87.72% | motif file (matrix) |
| 16 | 0.606 |  | 1e-21 | -50.143254 | 46.33% | 38.95% | motif file (matrix) |
| 17 | 0.708 |  | 1e-21 | -48.364759 | 81.09% | 74.93% | motif file (matrix) |
| 18 | 0.623 |  | 1e-20 | -47.180969 | 55.80% | 48.56% | motif file (matrix) |
| 19 | 0.609 |  | 1e-19 | -45.134718 | 52.41% | 45.34% | motif file (matrix) |
| 20 | 0.780 |  | 1e-17 | -39.900478 | 51.94% | 45.32% | motif file (matrix) |
| 21 | 0.606 |  | 1e-17 | -39.384774 | 94.07% | 90.46% | motif file (matrix) |
| 22 | 0.629 |  | 1e-15 | -35.020355 | 49.51% | 43.35% | motif file (matrix) |
| 23 | 0.741 |  | 1e-11 | -25.958146 | 64.51% | 59.39% | motif file (matrix) |
| 24 | 0.680 |  | 1e-9 | -22.276073 | 17.88% | 14.38% | motif file (matrix) |
| 25 | 0.712 |  | 1e-7 | -18.151638 | 37.09% | 33.00% | motif file (matrix) |