



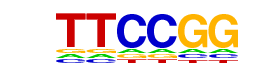





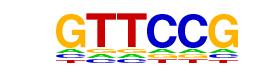

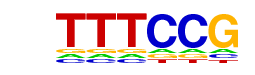


| p-value: | 1e-61 |
| log p-value: | -1.412e+02 |
| Information Content per bp: | 1.866 |
| Number of Target Sequences with motif | 2198.0 |
| Percentage of Target Sequences with motif | 46.91% |
| Number of Background Sequences with motif | 13909.6 |
| Percentage of Background Sequences with motif | 34.44% |
| Average Position of motif in Targets | 1008.5 +/- 344.8bp |
| Average Position of motif in Background | 1030.4 +/- 463.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.51 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.864 |  | 1e-54 | -125.293678 | 71.36% | 59.82% | motif file (matrix) |
| 2 | 0.946 |  | 1e-51 | -119.041907 | 45.77% | 34.37% | motif file (matrix) |
| 3 | 0.923 |  | 1e-47 | -109.765884 | 70.44% | 59.63% | motif file (matrix) |
| 4 | 0.864 |  | 1e-41 | -94.906576 | 93.00% | 86.48% | motif file (matrix) |
| 5 | 0.900 |  | 1e-40 | -92.115077 | 65.22% | 55.11% | motif file (matrix) |
| 6 | 0.642 |  | 1e-26 | -60.643831 | 55.89% | 47.62% | motif file (matrix) |
| 7 | 0.674 |  | 1e-25 | -58.269705 | 36.92% | 29.36% | motif file (matrix) |
| 8 | 0.687 |  | 1e-24 | -57.166291 | 56.55% | 48.54% | motif file (matrix) |
| 9 | 0.775 |  | 1e-24 | -56.604120 | 59.99% | 52.05% | motif file (matrix) |
| 10 | 0.609 |  | 1e-21 | -49.418264 | 95.03% | 91.15% | motif file (matrix) |
| 11 | 0.753 |  | 1e-18 | -42.918413 | 45.82% | 39.03% | motif file (matrix) |
| 12 | 0.648 |  | 1e-10 | -23.801650 | 61.29% | 56.35% | motif file (matrix) |
| 13 | 0.745 |  | 1e-9 | -22.733689 | 52.16% | 47.27% | motif file (matrix) |