






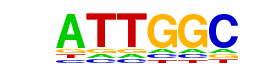





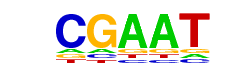
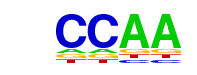



| p-value: | 1e-35 |
| log p-value: | -8.089e+01 |
| Information Content per bp: | 1.803 |
| Number of Target Sequences with motif | 2057.0 |
| Percentage of Target Sequences with motif | 43.90% |
| Number of Background Sequences with motif | 13965.5 |
| Percentage of Background Sequences with motif | 34.57% |
| Average Position of motif in Targets | 962.0 +/- 405.0bp |
| Average Position of motif in Background | 970.4 +/- 529.8bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.55 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.755 |  | 1e-31 | -71.771064 | 38.33% | 29.84% | motif file (matrix) |
| 2 | 0.918 |  | 1e-26 | -61.299816 | 54.61% | 46.29% | motif file (matrix) |
| 3 | 0.789 |  | 1e-23 | -54.707808 | 52.67% | 44.84% | motif file (matrix) |
| 4 | 0.667 |  | 1e-22 | -52.715690 | 47.18% | 39.59% | motif file (matrix) |
| 5 | 0.766 |  | 1e-20 | -48.117484 | 61.65% | 54.40% | motif file (matrix) |
| 6 | 0.737 |  | 1e-20 | -46.963533 | 50.00% | 42.80% | motif file (matrix) |
| 7 | 0.798 |  | 1e-18 | -42.934169 | 29.09% | 23.10% | motif file (matrix) |
| 8 | 0.786 |  | 1e-17 | -41.324994 | 47.42% | 40.73% | motif file (matrix) |
| 9 | 0.769 |  | 1e-17 | -39.476853 | 36.36% | 30.19% | motif file (matrix) |
| 10 | 0.657 |  | 1e-16 | -38.919310 | 95.80% | 92.65% | motif file (matrix) |
| 11 | 0.894 |  | 1e-15 | -34.939919 | 50.38% | 44.22% | motif file (matrix) |
| 12 | 0.617 |  | 1e-14 | -32.394312 | 66.05% | 60.31% | motif file (matrix) |
| 13 | 0.862 |  | 1e-9 | -22.560074 | 84.36% | 80.62% | motif file (matrix) |
| 14 | 0.629 |  | 1e-7 | -17.285828 | 58.62% | 54.47% | motif file (matrix) |
| 15 | 0.770 |  | 1e-1 | -2.395919 | 99.98% | 99.91% | motif file (matrix) |
| 16 | 0.641 |  | 1e0 | -1.007373 | 99.89% | 99.86% | motif file (matrix) |