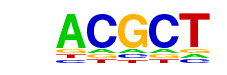

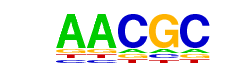





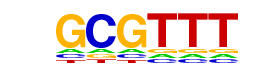


| p-value: | 1e-33 |
| log p-value: | -7.758e+01 |
| Information Content per bp: | 1.811 |
| Number of Target Sequences with motif | 3280.0 |
| Percentage of Target Sequences with motif | 70.00% |
| Number of Background Sequences with motif | 24635.2 |
| Percentage of Background Sequences with motif | 60.99% |
| Average Position of motif in Targets | 1027.3 +/- 497.0bp |
| Average Position of motif in Background | 1034.7 +/- 617.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.66 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.915 |  | 1e-20 | -47.874714 | 82.76% | 76.81% | motif file (matrix) |
| 2 | 0.683 |  | 1e-19 | -43.892760 | 31.88% | 25.63% | motif file (matrix) |
| 3 | 0.746 |  | 1e-18 | -41.552145 | 80.88% | 75.20% | motif file (matrix) |
| 4 | 0.667 |  | 1e-17 | -41.391448 | 62.72% | 56.05% | motif file (matrix) |
| 5 | 0.925 |  | 1e-15 | -36.541331 | 66.15% | 60.02% | motif file (matrix) |
| 6 | 0.746 |  | 1e-14 | -33.942953 | 55.53% | 49.45% | motif file (matrix) |
| 7 | 0.830 |  | 1e-14 | -33.006051 | 17.93% | 13.63% | motif file (matrix) |
| 8 | 0.705 |  | 1e-9 | -20.961157 | 20.55% | 16.94% | motif file (matrix) |
| 9 | 0.681 |  | 1e-9 | -20.825691 | 42.15% | 37.60% | motif file (matrix) |