









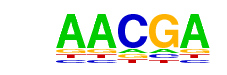
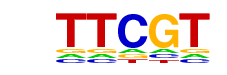




| p-value: | 1e-32 |
| log p-value: | -7.534e+01 |
| Information Content per bp: | 1.935 |
| Number of Target Sequences with motif | 3625.0 |
| Percentage of Target Sequences with motif | 77.36% |
| Number of Background Sequences with motif | 27894.5 |
| Percentage of Background Sequences with motif | 69.06% |
| Average Position of motif in Targets | 1052.2 +/- 500.1bp |
| Average Position of motif in Background | 1059.5 +/- 621.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.41 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.901 |  | 1e-28 | -66.616416 | 64.32% | 55.77% | motif file (matrix) |
| 2 | 0.710 |  | 1e-28 | -65.755231 | 74.93% | 67.02% | motif file (matrix) |
| 3 | 0.644 |  | 1e-21 | -49.479080 | 36.53% | 29.59% | motif file (matrix) |
| 4 | 0.678 |  | 1e-17 | -40.145508 | 59.41% | 52.80% | motif file (matrix) |
| 5 | 0.666 |  | 1e-17 | -39.313152 | 69.48% | 63.23% | motif file (matrix) |
| 6 | 0.622 |  | 1e-16 | -39.118113 | 65.54% | 59.15% | motif file (matrix) |
| 7 | 0.662 |  | 1e-16 | -39.031614 | 62.27% | 55.81% | motif file (matrix) |
| 8 | 0.647 |  | 1e-16 | -37.735211 | 74.73% | 68.89% | motif file (matrix) |
| 9 | 0.612 |  | 1e-14 | -33.606976 | 92.19% | 88.54% | motif file (matrix) |
| 10 | 0.648 |  | 1e-14 | -32.638389 | 51.66% | 45.72% | motif file (matrix) |
| 11 | 0.672 |  | 1e-10 | -24.038371 | 67.26% | 62.43% | motif file (matrix) |
| 12 | 0.696 |  | 1e-8 | -19.951517 | 68.35% | 64.04% | motif file (matrix) |
| 13 | 0.613 |  | 1e-7 | -16.680103 | 32.80% | 29.03% | motif file (matrix) |
| 14 | 0.761 |  | 1e-1 | -3.293997 | 100.00% | 99.93% | motif file (matrix) |