










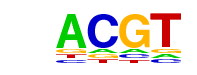









| p-value: | 1e-168 |
| log p-value: | -3.884e+02 |
| Information Content per bp: | 1.813 |
| Number of Target Sequences with motif | 13108.0 |
| Percentage of Target Sequences with motif | 49.62% |
| Number of Background Sequences with motif | 10497.5 |
| Percentage of Background Sequences with motif | 41.16% |
| Average Position of motif in Targets | 200.6 +/- 113.3bp |
| Average Position of motif in Background | 199.9 +/- 138.4bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.53 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.817 |  | 1e-150 | -347.258757 | 62.53% | 54.56% | motif file (matrix) |
| 2 | 0.796 |  | 1e-145 | -335.899005 | 57.30% | 49.39% | motif file (matrix) |
| 3 | 0.663 |  | 1e-144 | -332.814225 | 29.78% | 22.94% | motif file (matrix) |
| 4 | 0.847 |  | 1e-138 | -319.892499 | 59.32% | 51.63% | motif file (matrix) |
| 5 | 0.798 |  | 1e-129 | -299.122460 | 72.26% | 65.27% | motif file (matrix) |
| 6 | 0.909 |  | 1e-118 | -273.483973 | 59.12% | 52.01% | motif file (matrix) |
| 7 | 0.722 |  | 1e-117 | -270.361519 | 49.84% | 42.78% | motif file (matrix) |
| 8 | 0.604 |  | 1e-107 | -248.601872 | 40.78% | 34.24% | motif file (matrix) |
| 9 | 0.767 |  | 1e-98 | -226.765488 | 46.60% | 40.18% | motif file (matrix) |
| 10 | 0.665 |  | 1e-95 | -219.359957 | 66.24% | 60.05% | motif file (matrix) |
| 11 | 0.642 |  | 1e-95 | -219.093177 | 40.18% | 34.06% | motif file (matrix) |
| 12 | 0.667 |  | 1e-79 | -183.245494 | 41.46% | 35.81% | motif file (matrix) |
| 13 | 0.808 |  | 1e-79 | -183.245494 | 41.46% | 35.81% | motif file (matrix) |
| 14 | 0.703 |  | 1e-64 | -149.311415 | 55.27% | 50.02% | motif file (matrix) |
| 15 | 0.803 |  | 1e-63 | -146.795801 | 72.39% | 67.58% | motif file (matrix) |
| 16 | 0.654 |  | 1e-62 | -144.039757 | 45.83% | 40.74% | motif file (matrix) |
| 17 | 0.654 |  | 1e-62 | -143.276397 | 63.27% | 58.23% | motif file (matrix) |
| 18 | 0.631 |  | 1e-60 | -138.574662 | 12.32% | 9.27% | motif file (matrix) |
| 19 | 0.768 |  | 1e-44 | -102.141100 | 67.16% | 63.03% | motif file (matrix) |
| 20 | 0.693 |  | 1e-41 | -95.755602 | 17.62% | 14.59% | motif file (matrix) |