| p-value: | 1e-136 |
| log p-value: | -3.149e+02 |
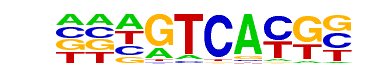
| Information Content per bp: | 1.733 |
| Number of Target Sequences with motif | 5436.0 |
| Percentage of Target Sequences with motif | 20.58% |
| Number of Background Sequences with motif | 3790.5 |
| Percentage of Background Sequences with motif | 14.86% |
| Average Position of motif in Targets | 198.4 +/- 115.2bp |
| Average Position of motif in Background | 196.4 +/- 123.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.14 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Atf1/MA0604.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.89
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TRCGTCA-
TACGTCAT |
|


|
|
MF0002.1_bZIP_CREB/G-box-like_subclass/Jaspar
| Match Rank: | 2 |
| Score: | 0.85
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TRCGTCA
-ACGTCA |
|


|
|
CREB1/MA0018.2/Jaspar
| Match Rank: | 3 |
| Score: | 0.82
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TRCGTCA
TGACGTCA |
|


|
|
Atf3/MA0605.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.81
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TRCGTCA--
-ACGTCATC |
|


|
|
Crem/MA0609.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.81
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TRCGTCA--
TTACGTCATN |
|


|
|
FOS::JUN/MA0099.2/Jaspar
| Match Rank: | 6 |
| Score: | 0.79
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TRCGTCA
TGAGTCA |
|


|
|
MAFG::NFE2L1/MA0089.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.77
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TRCGTCA--
---GTCATN |
|


|
|
Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 8 |
| Score: | 0.76
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TRCGTCA--
TGACGTCATC |
|


|
|
AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 9 |
| Score: | 0.75
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TRCGTCA-
GATGAGTCAT |
|


|
|
Pax2/MA0067.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.74
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | TRCGTCA---
--AGTCACGC |
|

|
|





















