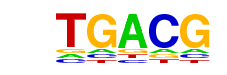






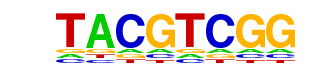


| p-value: | 1e-136 |
| log p-value: | -3.149e+02 |
| Information Content per bp: | 1.733 |
| Number of Target Sequences with motif | 5436.0 |
| Percentage of Target Sequences with motif | 20.58% |
| Number of Background Sequences with motif | 3790.5 |
| Percentage of Background Sequences with motif | 14.86% |
| Average Position of motif in Targets | 198.4 +/- 115.2bp |
| Average Position of motif in Background | 196.4 +/- 123.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.14 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.882 |  | 1e-89 | -205.920019 | 26.57% | 21.36% | motif file (matrix) |
| 2 | 0.610 |  | 1e-77 | -178.101689 | 26.53% | 21.68% | motif file (matrix) |
| 3 | 0.832 |  | 1e-73 | -170.390018 | 6.02% | 3.71% | motif file (matrix) |
| 4 | 0.644 |  | 1e-64 | -147.744010 | 25.34% | 20.99% | motif file (matrix) |
| 5 | 0.757 |  | 1e-49 | -114.777205 | 9.17% | 6.75% | motif file (matrix) |
| 6 | 0.741 |  | 1e-46 | -107.730584 | 91.80% | 89.14% | motif file (matrix) |
| 7 | 0.748 |  | 1e-45 | -104.950534 | 44.87% | 40.55% | motif file (matrix) |
| 8 | 0.694 |  | 1e-3 | -8.642866 | 0.17% | 0.10% | motif file (matrix) |