
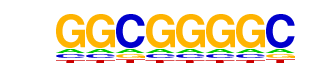
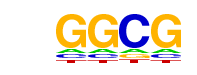



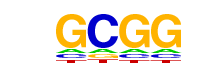
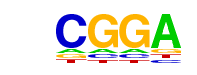








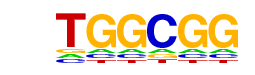


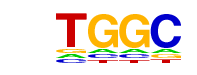


| p-value: | 1e-38 |
| log p-value: | -8.805e+01 |
| Information Content per bp: | 1.778 |
| Number of Target Sequences with motif | 6490.0 |
| Percentage of Target Sequences with motif | 98.05% |
| Number of Background Sequences with motif | 36010.9 |
| Percentage of Background Sequences with motif | 94.70% |
| Average Position of motif in Targets | 1082.9 +/- 413.7bp |
| Average Position of motif in Background | 1073.2 +/- 567.4bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 8.37 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.811 |  | 1e-22 | -52.545938 | 79.09% | 73.40% | motif file (matrix) |
| 2 | 0.758 |  | 1e-21 | -49.822250 | 97.08% | 94.41% | motif file (matrix) |
| 3 | 0.879 |  | 1e-18 | -42.376147 | 99.21% | 97.70% | motif file (matrix) |
| 4 | 0.640 |  | 1e-17 | -40.765308 | 62.15% | 56.45% | motif file (matrix) |
| 5 | 0.790 |  | 1e-16 | -37.636134 | 33.92% | 28.80% | motif file (matrix) |
| 6 | 0.707 |  | 1e-15 | -35.495438 | 58.26% | 52.91% | motif file (matrix) |
| 7 | 0.865 |  | 1e-14 | -34.454651 | 99.21% | 97.90% | motif file (matrix) |
| 8 | 0.670 |  | 1e-14 | -33.270919 | 99.38% | 98.20% | motif file (matrix) |
| 9 | 0.709 |  | 1e-13 | -31.182767 | 63.91% | 59.02% | motif file (matrix) |
| 10 | 0.656 |  | 1e-10 | -24.918252 | 48.03% | 43.62% | motif file (matrix) |
| 11 | 0.718 |  | 1e-10 | -24.804297 | 48.09% | 43.68% | motif file (matrix) |
| 12 | 0.641 |  | 1e-9 | -21.460327 | 44.69% | 40.66% | motif file (matrix) |
| 13 | 0.640 |  | 1e-9 | -21.025814 | 50.64% | 46.61% | motif file (matrix) |
| 14 | 0.811 |  | 1e-8 | -19.599993 | 54.39% | 50.51% | motif file (matrix) |
| 15 | 0.617 |  | 1e-5 | -11.926837 | 98.37% | 97.52% | motif file (matrix) |
| 16 | 0.876 |  | 1e-3 | -7.742996 | 99.94% | 99.74% | motif file (matrix) |
| 17 | 0.876 |  | 1e-2 | -6.295788 | 99.95% | 99.80% | motif file (matrix) |
| 18 | 0.631 |  | 1e-2 | -5.957957 | 99.67% | 99.39% | motif file (matrix) |
| 19 | 0.718 |  | 1e-2 | -5.017924 | 99.94% | 99.80% | motif file (matrix) |
| 20 | 0.684 |  | 1e0 | -1.189359 | 99.92% | 99.89% | motif file (matrix) |