ZBED1/MA0749.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.85
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TCGCGATA-
TATGTCGCGATAG |
|


|
|
GFX(?)/Promoter/Homer
| Match Rank: | 2 |
| Score: | 0.79
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TCGCGATA--
TCTCGCGAGAAT |
|


|
|
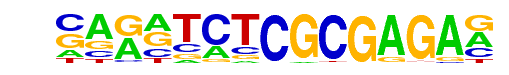
ZBTB33/MA0527.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.78
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------TCGCGATA-
NAGNTCTCGCGAGAN |
|

|
|
ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 4 |
| Score: | 0.77
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TCGCGATA--
GGNTCTCGCGAGAAC |
|


|
|
CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 5 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TCGCGATA--
TTGCAACATN |
|


|
|
E2F(E2F)/Hela-CellCycle-Expression/Homer
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TCGCGATA-
TTCGCGCGAAAA |
|


|
|
PB0139.1_Irf5_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TCGCGATA--
NNAATTCTCGNTNAN |
|


|
|
YY2/MA0748.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TCGCGATA-
GTCCGCCATTA |
|


|
|
PB0140.1_Irf6_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TCGCGATA--
ACCACTCTCGGTCAC |
|


|
|
PB0138.1_Irf4_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TCGCGATA--
AGTATTCTCGGTTGC |
|


|
|





















