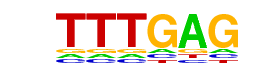
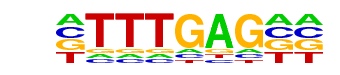
Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTTGAG--
CCTTTGATGT |
|

|
|
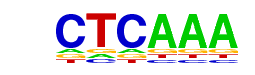
PB0082.1_Tcf3_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------TTTGAG-----
NNTTCCTTTGATCTANA |
|

|
|
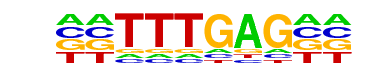
CHR(?)/Hela-CellCycle-Expression/Homer
| Match Rank: | 3 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTTGAG----
TTTGAAACCG |
|


|
|
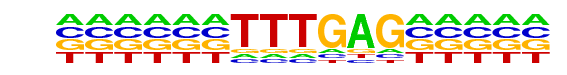
Nkx2-5(var.2)/MA0503.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTTGAG-----
CTTGAGTGGCT |
|


|
|
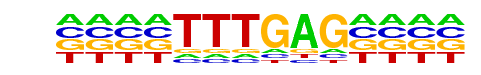
TCF7L2/MA0523.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.69
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTTGAG----
TNCCTTTGATCTTN |
|

|
|
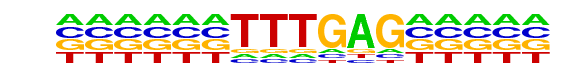
PB0084.1_Tcf7l2_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.68
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------TTTGAG-----
ATTTCCTTTGATCTATA |
|

|
|
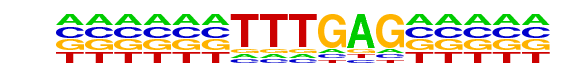
PB0083.1_Tcf7_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.68
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------TTTGAG-----
NNTTCCTTTGATCTNNA |
|

|
|
Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer
| Match Rank: | 8 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TTTGAG--
TNCCTTTGATGT |
|


|
|
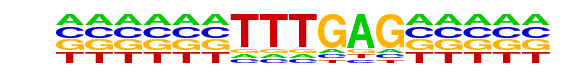
PB0040.1_Lef1_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.67
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------TTTGAG-----
AATCCCTTTGATCTATC |
|

|
|
LIN54/MA0619.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.67
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TTTGAG--
ATTTGAATT |
|

|
|