

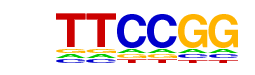



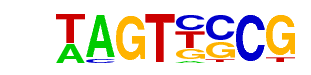







| p-value: | 1e-33 |
| log p-value: | -7.778e+01 |
| Information Content per bp: | 1.801 |
| Number of Target Sequences with motif | 3394.0 |
| Percentage of Target Sequences with motif | 51.28% |
| Number of Background Sequences with motif | 16417.1 |
| Percentage of Background Sequences with motif | 43.17% |
| Average Position of motif in Targets | 1021.2 +/- 420.3bp |
| Average Position of motif in Background | 1034.9 +/- 557.5bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.53 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.747 |  | 1e-33 | -76.650287 | 59.62% | 51.59% | motif file (matrix) |
| 2 | 0.929 |  | 1e-27 | -63.270274 | 67.20% | 60.15% | motif file (matrix) |
| 3 | 0.785 |  | 1e-25 | -59.793573 | 62.67% | 55.68% | motif file (matrix) |
| 4 | 0.860 |  | 1e-25 | -59.393287 | 91.52% | 87.07% | motif file (matrix) |
| 5 | 0.621 |  | 1e-25 | -58.488329 | 56.35% | 49.36% | motif file (matrix) |
| 6 | 0.639 |  | 1e-13 | -31.196376 | 48.57% | 43.58% | motif file (matrix) |
| 7 | 0.685 |  | 1e-13 | -29.971714 | 22.83% | 18.88% | motif file (matrix) |
| 8 | 0.644 |  | 1e-11 | -26.551549 | 32.39% | 28.19% | motif file (matrix) |
| 9 | 0.620 |  | 1e-11 | -25.886346 | 53.59% | 49.06% | motif file (matrix) |
| 10 | 0.662 |  | 1e-4 | -10.906531 | 59.12% | 56.39% | motif file (matrix) |
| 11 | 0.622 |  | 1e0 | -1.309991 | 97.75% | 97.61% | motif file (matrix) |
| 12 | 0.756 |  | 1e0 | -1.060148 | 99.97% | 99.95% | motif file (matrix) |