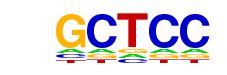
| p-value: | 1e-2 |
| log p-value: | -6.453e+00 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 6569.0 |
| Percentage of Target Sequences with motif | 99.24% |
| Number of Background Sequences with motif | 37584.8 |
| Percentage of Background Sequences with motif | 98.84% |
| Average Position of motif in Targets | 1076.0 +/- 479.3bp |
| Average Position of motif in Background | 1065.4 +/- 603.6bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 7.76 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
POL013.1_MED-1/Jaspar
| Match Rank: | 1 |
| Score: | 0.96
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GGAGC
CGGAGC |
|


|
|
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 2 |
| Score: | 0.79
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GGAGC-
-CAGCC |
|


|
|
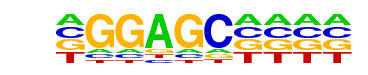
Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 3 |
| Score: | 0.78
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GGAGC----
GGGAGGACNG |
|

|
|
ZNF415(Zf)/HEK293-ZNF415.GFP-ChIP-Seq(GSE58341)/Homer
| Match Rank: | 4 |
| Score: | 0.74
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------GGAGC-
GRTGMTRGAGCC |
|


|
|
POL008.1_DCE_S_I/Jaspar
| Match Rank: | 5 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GGAGC
NGAAGC |
|


|
|
PB0099.1_Zfp691_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.67
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GGAGC--------
NNNNTGAGCACTGTNNG |
|


|
|
PB0154.1_Osr1_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | -7
| | Orientation: | reverse strand |
| Alignment: | -------GGAGC----
NNNTTAGGTAGCNTNT |
|


|
|
PB0203.1_Zfp691_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------GGAGC------
NTNNNAGGAGTCTCNTN |
|


|
|
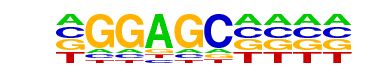
ZNF189(Zf)/HEK293-ZNF189.GFP-ChIP-Seq(GSE58341)/Homer
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGAGC----
TGGAACAGMA |
|

|
|
PB0050.1_Osr1_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.61
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------GGAGC----
ATTTACAGTAGCAAAA |
|


|
|