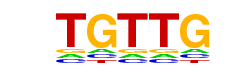
| p-value: | 1e-2 |
| log p-value: | -5.233e+00 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 5733.0 |
| Percentage of Target Sequences with motif | 86.61% |
| Number of Background Sequences with motif | 32480.3 |
| Percentage of Background Sequences with motif | 85.42% |
| Average Position of motif in Targets | 1036.3 +/- 608.3bp |
| Average Position of motif in Background | 1022.7 +/- 700.8bp |
| Strand Bias (log2 ratio + to - strand density) | -0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.75 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0120.1_Foxj1_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.76
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------CAACA----
ATGTCACAACAACAC |
|


|
|
SCRT2/MA0744.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.74
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CAACA-----
ATGCAACAGGTGG |
|


|
|
PB0122.1_Foxk1_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.74
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------CAACA---
CAAACAACAACACCT |
|


|
|
SCRT1/MA0743.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.74
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CAACA-------
GAGCAACAGGTGGTT |
|


|
|
MF0011.1_HMG_class/Jaspar
| Match Rank: | 5 |
| Score: | 0.74
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CAACA--
-AACAAT |
|


|
|
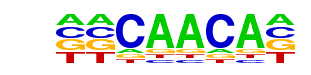
Foxj2/MA0614.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.72
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CAACA-
GTAAACAA |
|

|
|
PB0121.1_Foxj3_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.72
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------CAACA-----
AACACCAAAACAAAGGA |
|


|
|
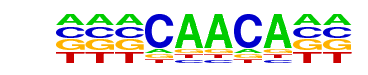
CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 8 |
| Score: | 0.71
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CAACA--
TTGCAACATN |
|

|
|
FOXO4/MA0848.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.70
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CAACA
GTAAACA |
|


|
|
Sox5/MA0087.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.70
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CAACA--
NAACAAT |
|


|
|