





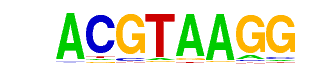







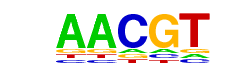




| p-value: | 1e-22 |
| log p-value: | -5.145e+01 |
| Information Content per bp: | 1.877 |
| Number of Target Sequences with motif | 4126.0 |
| Percentage of Target Sequences with motif | 62.34% |
| Number of Background Sequences with motif | 21248.0 |
| Percentage of Background Sequences with motif | 55.88% |
| Average Position of motif in Targets | 1026.8 +/- 561.9bp |
| Average Position of motif in Background | 1025.9 +/- 700.4bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.72 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.905 |  | 1e-21 | -49.431212 | 91.99% | 88.08% | motif file (matrix) |
| 2 | 0.875 |  | 1e-19 | -45.389305 | 86.57% | 82.05% | motif file (matrix) |
| 3 | 0.710 |  | 1e-17 | -40.222394 | 48.81% | 43.09% | motif file (matrix) |
| 4 | 0.840 |  | 1e-16 | -37.912298 | 39.30% | 33.95% | motif file (matrix) |
| 5 | 0.665 |  | 1e-16 | -37.452473 | 68.92% | 63.67% | motif file (matrix) |
| 6 | 0.678 |  | 1e-14 | -33.153832 | 49.21% | 44.04% | motif file (matrix) |
| 7 | 0.680 |  | 1e-14 | -32.749789 | 40.53% | 35.55% | motif file (matrix) |
| 8 | 0.691 |  | 1e-13 | -31.582781 | 96.62% | 94.48% | motif file (matrix) |
| 9 | 0.638 |  | 1e-12 | -28.402628 | 33.69% | 29.29% | motif file (matrix) |
| 10 | 0.673 |  | 1e-11 | -26.229736 | 14.07% | 11.08% | motif file (matrix) |
| 11 | 0.763 |  | 1e-10 | -23.262998 | 38.95% | 34.84% | motif file (matrix) |
| 12 | 0.742 |  | 1e-9 | -21.828027 | 46.26% | 42.17% | motif file (matrix) |
| 13 | 0.607 |  | 1e-8 | -18.864441 | 69.13% | 65.56% | motif file (matrix) |
| 14 | 0.603 |  | 1e-8 | -18.646140 | 70.49% | 66.98% | motif file (matrix) |
| 15 | 0.607 |  | 1e-7 | -17.238897 | 61.58% | 58.04% | motif file (matrix) |
| 16 | 0.813 |  | 1e-1 | -2.593298 | 99.98% | 99.93% | motif file (matrix) |
| 17 | 0.809 |  | 1e-1 | -2.335213 | 99.98% | 99.93% | motif file (matrix) |