




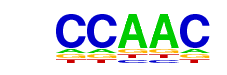
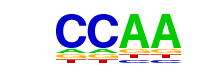

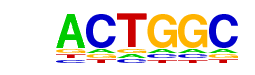


| p-value: | 1e-17 |
| log p-value: | -4.099e+01 |
| Information Content per bp: | 1.790 |
| Number of Target Sequences with motif | 1330.0 |
| Percentage of Target Sequences with motif | 20.09% |
| Number of Background Sequences with motif | 5969.4 |
| Percentage of Background Sequences with motif | 15.70% |
| Average Position of motif in Targets | 996.6 +/- 532.0bp |
| Average Position of motif in Background | 1017.1 +/- 649.8bp |
| Strand Bias (log2 ratio + to - strand density) | 0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.13 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.639 |  | 1e-16 | -37.195308 | 42.24% | 36.87% | motif file (matrix) |
| 2 | 0.761 |  | 1e-14 | -33.407234 | 45.93% | 40.78% | motif file (matrix) |
| 3 | 0.711 |  | 1e-9 | -21.833552 | 61.43% | 57.37% | motif file (matrix) |
| 4 | 0.827 |  | 1e-8 | -19.826972 | 82.40% | 79.30% | motif file (matrix) |
| 5 | 0.727 |  | 1e-2 | -5.317633 | 36.80% | 35.15% | motif file (matrix) |
| 6 | 0.606 |  | 1e-2 | -5.109523 | 92.88% | 91.98% | motif file (matrix) |
| 7 | 0.746 |  | 1e-1 | -2.506585 | 99.97% | 99.91% | motif file (matrix) |
| 8 | 0.707 |  | 1e0 | -1.897041 | 99.95% | 99.90% | motif file (matrix) |
| 9 | 0.725 |  | 1e0 | -1.148437 | 99.94% | 99.91% | motif file (matrix) |