| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-48 | -1.108e+02 | 0.0000 | 3242.0 | 69.18% | 23524.5 | 58.24% | motif file (matrix) |
pdf |
| 2 |  | ETS(ETS)/Promoter/Homer | 1e-47 | -1.097e+02 | 0.0000 | 2166.0 | 46.22% | 14237.2 | 35.25% | motif file (matrix) |
pdf |
| 3 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-47 | -1.088e+02 | 0.0000 | 3190.0 | 68.08% | 23092.1 | 57.17% | motif file (matrix) |
pdf |
| 4 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-39 | -9.103e+01 | 0.0000 | 2935.0 | 62.63% | 21207.2 | 52.50% | motif file (matrix) |
pdf |
| 5 |  | NFY(CCAAT)/Promoter/Homer | 1e-31 | -7.252e+01 | 0.0000 | 2577.0 | 54.99% | 18547.2 | 45.92% | motif file (matrix) |
pdf |
| 6 | 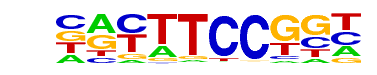 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-29 | -6.907e+01 | 0.0000 | 3778.0 | 80.62% | 29507.2 | 73.05% | motif file (matrix) |
pdf |
| 7 |  | CRE(bZIP)/Promoter/Homer | 1e-27 | -6.415e+01 | 0.0000 | 1256.0 | 26.80% | 7965.9 | 19.72% | motif file (matrix) |
pdf |
| 8 |  | GFY-Staf(?,Zf)/Promoter/Homer | 1e-24 | -5.612e+01 | 0.0000 | 629.0 | 13.42% | 3473.3 | 8.60% | motif file (matrix) |
pdf |
| 9 |  | Sp1(Zf)/Promoter/Homer | 1e-23 | -5.517e+01 | 0.0000 | 2638.0 | 56.30% | 19561.8 | 48.43% | motif file (matrix) |
pdf |
| 10 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-23 | -5.371e+01 | 0.0000 | 3231.0 | 68.95% | 24846.1 | 61.51% | motif file (matrix) |
pdf |
| 11 |  | YY1(Zf)/Promoter/Homer | 1e-23 | -5.332e+01 | 0.0000 | 717.0 | 15.30% | 4149.0 | 10.27% | motif file (matrix) |
pdf |
| 12 |  | JunD(bZIP)/K562-JunD-ChIP-Seq/Homer | 1e-22 | -5.110e+01 | 0.0000 | 448.0 | 9.56% | 2300.3 | 5.69% | motif file (matrix) |
pdf |
| 13 |  | Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer | 1e-21 | -4.982e+01 | 0.0000 | 1921.0 | 40.99% | 13659.6 | 33.82% | motif file (matrix) |
pdf |
| 14 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-20 | -4.823e+01 | 0.0000 | 3099.0 | 66.13% | 23832.1 | 59.00% | motif file (matrix) |
pdf |
| 15 | 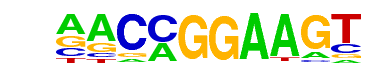 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-20 | -4.757e+01 | 0.0000 | 3804.0 | 81.18% | 30328.5 | 75.08% | motif file (matrix) |
pdf |
| 16 |  | GFY(?)/Promoter/Homer | 1e-18 | -4.325e+01 | 0.0000 | 650.0 | 13.87% | 3849.7 | 9.53% | motif file (matrix) |
pdf |
| 17 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-17 | -4.114e+01 | 0.0000 | 3291.0 | 70.23% | 25792.3 | 63.85% | motif file (matrix) |
pdf |
| 18 |  | Atf7(bZIP)/3T3L1-Atf7-ChIP-Seq(GSE56872)/Homer | 1e-15 | -3.675e+01 | 0.0000 | 1364.0 | 29.11% | 9521.6 | 23.57% | motif file (matrix) |
pdf |
| 19 |  | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-15 | -3.550e+01 | 0.0000 | 2410.0 | 51.43% | 18261.5 | 45.21% | motif file (matrix) |
pdf |
| 20 |  | Atf2(bZIP)/3T3L1-Atf2-ChIP-Seq(GSE56872)/Homer | 1e-14 | -3.390e+01 | 0.0000 | 1033.0 | 22.04% | 6979.4 | 17.28% | motif file (matrix) |
pdf |
| 21 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-14 | -3.381e+01 | 0.0000 | 2914.0 | 62.19% | 22702.9 | 56.21% | motif file (matrix) |
pdf |
| 22 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-13 | -3.149e+01 | 0.0000 | 852.0 | 18.18% | 5638.6 | 13.96% | motif file (matrix) |
pdf |
| 23 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-11 | -2.584e+01 | 0.0000 | 4109.0 | 87.69% | 33920.4 | 83.98% | motif file (matrix) |
pdf |
| 24 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-10 | -2.484e+01 | 0.0000 | 3262.0 | 69.61% | 26162.2 | 64.77% | motif file (matrix) |
pdf |
| 25 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-10 | -2.365e+01 | 0.0000 | 2096.0 | 44.73% | 16079.9 | 39.81% | motif file (matrix) |
pdf |
| 26 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-9 | -2.169e+01 | 0.0000 | 3781.0 | 80.69% | 31003.6 | 76.76% | motif file (matrix) |
pdf |
| 27 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-8 | -2.038e+01 | 0.0000 | 4237.0 | 90.42% | 35339.9 | 87.49% | motif file (matrix) |
pdf |
| 28 |  | KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer | 1e-8 | -1.912e+01 | 0.0000 | 4395.0 | 93.79% | 36927.2 | 91.42% | motif file (matrix) |
pdf |
| 29 |  | HIF-1a(bHLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-8 | -1.862e+01 | 0.0000 | 1181.0 | 25.20% | 8698.6 | 21.54% | motif file (matrix) |
pdf |
| 30 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-8 | -1.845e+01 | 0.0000 | 3310.0 | 70.64% | 26894.8 | 66.58% | motif file (matrix) |
pdf |
| 31 |  | Klf9(Zf)/GBM-Klf9-ChIP-Seq(GSE62211)/Homer | 1e-7 | -1.678e+01 | 0.0000 | 2256.0 | 48.14% | 17790.3 | 44.04% | motif file (matrix) |
pdf |
| 32 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-7 | -1.631e+01 | 0.0000 | 1293.0 | 27.59% | 9722.5 | 24.07% | motif file (matrix) |
pdf |
| 33 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-6 | -1.560e+01 | 0.0000 | 1186.0 | 25.31% | 8877.5 | 21.98% | motif file (matrix) |
pdf |
| 34 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-6 | -1.557e+01 | 0.0000 | 3033.0 | 64.72% | 24599.1 | 60.90% | motif file (matrix) |
pdf |
| 35 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-6 | -1.521e+01 | 0.0000 | 2004.0 | 42.77% | 15732.6 | 38.95% | motif file (matrix) |
pdf |
| 36 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-5 | -1.358e+01 | 0.0000 | 2818.0 | 60.14% | 22837.7 | 56.54% | motif file (matrix) |
pdf |
| 37 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-5 | -1.350e+01 | 0.0000 | 3539.0 | 75.52% | 29210.8 | 72.32% | motif file (matrix) |
pdf |
| 38 |  | E2F(E2F)/Hela-CellCycle-Expression/Homer | 1e-5 | -1.212e+01 | 0.0000 | 561.0 | 11.97% | 3987.4 | 9.87% | motif file (matrix) |
pdf |
| 39 |  | Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-5 | -1.203e+01 | 0.0000 | 4462.0 | 95.22% | 37819.0 | 93.63% | motif file (matrix) |
pdf |
| 40 |  | HIF2a(bHLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer | 1e-4 | -1.133e+01 | 0.0001 | 1560.0 | 33.29% | 12221.2 | 30.26% | motif file (matrix) |
pdf |
| 41 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-4 | -1.059e+01 | 0.0002 | 3663.0 | 78.17% | 30497.5 | 75.50% | motif file (matrix) |
pdf |
| 42 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-4 | -1.054e+01 | 0.0002 | 3773.0 | 80.52% | 31489.3 | 77.96% | motif file (matrix) |
pdf |
| 43 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-4 | -9.699e+00 | 0.0005 | 1963.0 | 41.89% | 15743.8 | 38.98% | motif file (matrix) |
pdf |
| 44 |  | Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer | 1e-4 | -9.622e+00 | 0.0005 | 4366.0 | 93.17% | 36988.4 | 91.57% | motif file (matrix) |
pdf |
| 45 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-4 | -9.293e+00 | 0.0007 | 1579.0 | 33.70% | 12519.4 | 30.99% | motif file (matrix) |
pdf |
| 46 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-3 | -8.833e+00 | 0.0010 | 483.0 | 10.31% | 3505.8 | 8.68% | motif file (matrix) |
pdf |
| 47 |  | PAX5(Paired,Homeobox),condensed/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-3 | -8.785e+00 | 0.0010 | 415.0 | 8.86% | 2967.1 | 7.35% | motif file (matrix) |
pdf |
| 48 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-3 | -8.248e+00 | 0.0017 | 2089.0 | 44.58% | 16932.4 | 41.92% | motif file (matrix) |
pdf |
| 49 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.212e+00 | 0.0018 | 4111.0 | 87.73% | 34695.1 | 85.89% | motif file (matrix) |
pdf |
| 50 |  | GFX(?)/Promoter/Homer | 1e-3 | -8.108e+00 | 0.0019 | 182.0 | 3.88% | 1185.9 | 2.94% | motif file (matrix) |
pdf |
| 51 |  | Nkx2.2(Homeobox)/NPC-Nkx2.2-ChIP-Seq(GSE61673)/Homer | 1e-3 | -7.829e+00 | 0.0025 | 3722.0 | 79.43% | 31212.8 | 77.27% | motif file (matrix) |
pdf |
| 52 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.462e+00 | 0.0035 | 3167.0 | 67.58% | 26334.1 | 65.20% | motif file (matrix) |
pdf |
| 53 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-3 | -7.410e+00 | 0.0036 | 3795.0 | 80.99% | 31895.5 | 78.96% | motif file (matrix) |
pdf |
| 54 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-3 | -7.311e+00 | 0.0039 | 3541.0 | 75.57% | 29642.4 | 73.39% | motif file (matrix) |
pdf |
| 55 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-3 | -7.233e+00 | 0.0042 | 3373.0 | 71.98% | 28164.5 | 69.73% | motif file (matrix) |
pdf |
| 56 |  | Nkx2.1(Homeobox)/LungAC-Nkx2.1-ChIP-Seq(GSE43252)/Homer | 1e-2 | -6.902e+00 | 0.0057 | 4170.0 | 88.99% | 35313.1 | 87.42% | motif file (matrix) |
pdf |
| 57 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-2 | -6.884e+00 | 0.0057 | 1324.0 | 28.25% | 10556.8 | 26.14% | motif file (matrix) |
pdf |
| 58 |  | Bapx1(Homeobox)/VertebralCol-Bapx1-ChIP-Seq(GSE36672)/Homer | 1e-2 | -6.860e+00 | 0.0058 | 3658.0 | 78.06% | 30716.1 | 76.04% | motif file (matrix) |
pdf |
| 59 |  | NRF(NRF)/Promoter/Homer | 1e-2 | -6.805e+00 | 0.0060 | 1318.0 | 28.13% | 10512.3 | 26.03% | motif file (matrix) |
pdf |
| 60 |  | bHLHE40(bHLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.437e+00 | 0.0085 | 1124.0 | 23.99% | 8914.0 | 22.07% | motif file (matrix) |
pdf |
| 61 |  | E-box(bHLH)/Promoter/Homer | 1e-2 | -6.404e+00 | 0.0087 | 462.0 | 9.86% | 3454.6 | 8.55% | motif file (matrix) |
pdf |
| 62 |  | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-2 | -6.248e+00 | 0.0100 | 458.0 | 9.77% | 3430.7 | 8.49% | motif file (matrix) |
pdf |
| 63 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-2 | -6.146e+00 | 0.0108 | 916.0 | 19.55% | 7201.3 | 17.83% | motif file (matrix) |
pdf |
| 64 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-2 | -5.953e+00 | 0.0129 | 1602.0 | 34.19% | 12986.9 | 32.15% | motif file (matrix) |
pdf |
| 65 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-2 | -5.900e+00 | 0.0134 | 3328.0 | 71.02% | 27886.8 | 69.04% | motif file (matrix) |
pdf |
| 66 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.808e+00 | 0.0145 | 4564.0 | 97.40% | 39039.1 | 96.65% | motif file (matrix) |
pdf |
| 67 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.742e+00 | 0.0153 | 992.0 | 21.17% | 7866.0 | 19.47% | motif file (matrix) |
pdf |
| 68 |  | GSC(Homeobox)/FrogEmbryos-GSC-ChIP-Seq(DRA000576)/Homer | 1e-2 | -5.709e+00 | 0.0156 | 2261.0 | 48.25% | 18640.8 | 46.15% | motif file (matrix) |
pdf |
| 69 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.944e+00 | 0.0330 | 1466.0 | 31.28% | 11931.1 | 29.54% | motif file (matrix) |
pdf |
| 70 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -4.862e+00 | 0.0353 | 1790.0 | 38.20% | 14696.1 | 36.38% | motif file (matrix) |
pdf |
| 71 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.818e+00 | 0.0363 | 1416.0 | 30.22% | 11520.5 | 28.52% | motif file (matrix) |
pdf |
| 72 |  | ETS:RUNX(ETS,Runt)/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -4.766e+00 | 0.0377 | 444.0 | 9.48% | 3402.4 | 8.42% | motif file (matrix) |
pdf |
| 73 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo_et_al.)/Homer | 1e-2 | -4.707e+00 | 0.0395 | 2296.0 | 49.00% | 19050.4 | 47.16% | motif file (matrix) |
pdf |
| 74 |  | Pbx3(Homeobox)/GM12878-PBX3-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.668e+00 | 0.0405 | 650.0 | 13.87% | 5105.5 | 12.64% | motif file (matrix) |
pdf |
| 75 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-2 | -4.648e+00 | 0.0407 | 1379.0 | 29.43% | 11224.1 | 27.79% | motif file (matrix) |
pdf |
| 76 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -4.638e+00 | 0.0407 | 2553.0 | 54.48% | 21274.6 | 52.67% | motif file (matrix) |
pdf |























































































































































